21. Xarray Interpolation, Groupby, Resample, Rolling, and Coarsen#
Attribution: This notebook is a revision of the Xarray Interpolation, Groupby, Resample, Rolling, and Coarsen notebook by Ryan Abernathey from An Introduction to Earth and Environmental Data Science. Thanks to Aiyin Zhang for preparing this notebook.
In this lesson, we cover some more advanced aspects of xarray.
You can access this notebook (in a Docker image) on this GitHub repo.
import numpy as np
import xarray as xr
from matplotlib import pyplot as plt
21.1. Interpolation#
In the previous lesson on xarray, we learned how to select data based on its dimension coordinates and align data with dimension different coordinates.
But what if we want to estimate the value of the data variables at different coordinates.
This is where interpolation comes in.
# we write it out explicitly so we can see each point.
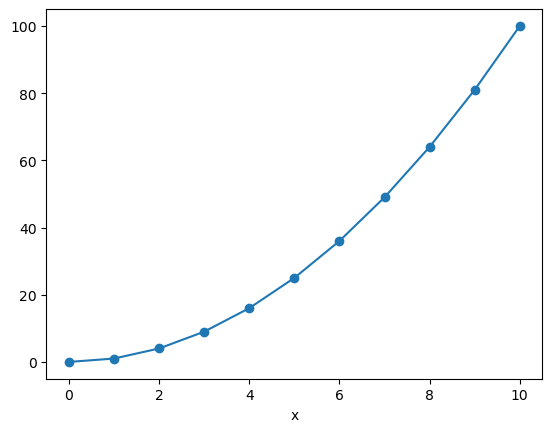
x_data = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10])
f = xr.DataArray(x_data**2, dims=['x'], coords={'x': x_data})
f
<xarray.DataArray (x: 11)> Size: 88B array([ 0, 1, 4, 9, 16, 25, 36, 49, 64, 81, 100]) Coordinates: * x (x) int64 88B 0 1 2 3 4 5 6 7 8 9 10
f.plot(marker='o')
[<matplotlib.lines.Line2D at 0xffff5c6420d0>]
f.sel(x=3)
<xarray.DataArray ()> Size: 8B
array(9)
Coordinates:
x int64 8B 3We only have data on the integer points in x. But what if we wanted to estimate the value at, say, 4.5?
f.sel(x=4.5)
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File /opt/conda/envs/xarray_tutorial/lib/python3.13/site-packages/pandas/core/indexes/base.py:3805, in Index.get_loc(self, key)
3804 try:
-> 3805 return self._engine.get_loc(casted_key)
3806 except KeyError as err:
File index.pyx:167, in pandas._libs.index.IndexEngine.get_loc()
File index.pyx:175, in pandas._libs.index.IndexEngine.get_loc()
File pandas/_libs/index_class_helper.pxi:70, in pandas._libs.index.Int64Engine._check_type()
KeyError: 4.5
The above exception was the direct cause of the following exception:
KeyError Traceback (most recent call last)
File /opt/conda/envs/xarray_tutorial/lib/python3.13/site-packages/xarray/core/indexes.py:789, in PandasIndex.sel(self, labels, method, tolerance)
788 try:
--> 789 indexer = self.index.get_loc(label_value)
790 except KeyError as e:
File /opt/conda/envs/xarray_tutorial/lib/python3.13/site-packages/pandas/core/indexes/base.py:3812, in Index.get_loc(self, key)
3811 raise InvalidIndexError(key)
-> 3812 raise KeyError(key) from err
3813 except TypeError:
3814 # If we have a listlike key, _check_indexing_error will raise
3815 # InvalidIndexError. Otherwise we fall through and re-raise
3816 # the TypeError.
KeyError: 4.5
The above exception was the direct cause of the following exception:
KeyError Traceback (most recent call last)
Cell In[5], line 1
----> 1 f.sel(x=4.5)
File /opt/conda/envs/xarray_tutorial/lib/python3.13/site-packages/xarray/core/dataarray.py:1675, in DataArray.sel(self, indexers, method, tolerance, drop, **indexers_kwargs)
1559 def sel(
1560 self,
1561 indexers: Mapping[Any, Any] | None = None,
(...)
1565 **indexers_kwargs: Any,
1566 ) -> Self:
1567 """Return a new DataArray whose data is given by selecting index
1568 labels along the specified dimension(s).
1569
(...)
1673 Dimensions without coordinates: points
1674 """
-> 1675 ds = self._to_temp_dataset().sel(
1676 indexers=indexers,
1677 drop=drop,
1678 method=method,
1679 tolerance=tolerance,
1680 **indexers_kwargs,
1681 )
1682 return self._from_temp_dataset(ds)
File /opt/conda/envs/xarray_tutorial/lib/python3.13/site-packages/xarray/core/dataset.py:3223, in Dataset.sel(self, indexers, method, tolerance, drop, **indexers_kwargs)
3155 """Returns a new dataset with each array indexed by tick labels
3156 along the specified dimension(s).
3157
(...)
3220
3221 """
3222 indexers = either_dict_or_kwargs(indexers, indexers_kwargs, "sel")
-> 3223 query_results = map_index_queries(
3224 self, indexers=indexers, method=method, tolerance=tolerance
3225 )
3227 if drop:
3228 no_scalar_variables = {}
File /opt/conda/envs/xarray_tutorial/lib/python3.13/site-packages/xarray/core/indexing.py:194, in map_index_queries(obj, indexers, method, tolerance, **indexers_kwargs)
192 results.append(IndexSelResult(labels))
193 else:
--> 194 results.append(index.sel(labels, **options))
196 merged = merge_sel_results(results)
198 # drop dimension coordinates found in dimension indexers
199 # (also drop multi-index if any)
200 # (.sel() already ensures alignment)
File /opt/conda/envs/xarray_tutorial/lib/python3.13/site-packages/xarray/core/indexes.py:791, in PandasIndex.sel(self, labels, method, tolerance)
789 indexer = self.index.get_loc(label_value)
790 except KeyError as e:
--> 791 raise KeyError(
792 f"not all values found in index {coord_name!r}. "
793 "Try setting the `method` keyword argument (example: method='nearest')."
794 ) from e
796 elif label_array.dtype.kind == "b":
797 indexer = label_array
KeyError: "not all values found in index 'x'. Try setting the `method` keyword argument (example: method='nearest')."
Interpolation to the rescue!
f.interp(x=4.5)
<xarray.DataArray ()> Size: 8B
array(20.5)
Coordinates:
x float64 8B 4.5Interpolation uses scipy.interpolate under the hood. There are different modes of interpolation.
f.interp(x=4.5, method='linear').values
array(20.5)
f.interp(x=4.5, method='nearest').values
array(16.)
f.interp(x=4.5, method='cubic').values
array(20.25)
We can interpolate to a whole new set of coordinates at once:
x_new = x_data + 0.5
f_interp_linear = f.interp(x=x_new, method='linear')
f_interp_cubic = f.interp(x=x_new, method='cubic')
f.plot(marker='o', label='original')
f_interp_linear.plot(marker='o', label='linear')
f_interp_cubic.plot(marker='o', label='cubic')
plt.legend()
<matplotlib.legend.Legend at 0xffff545501a0>
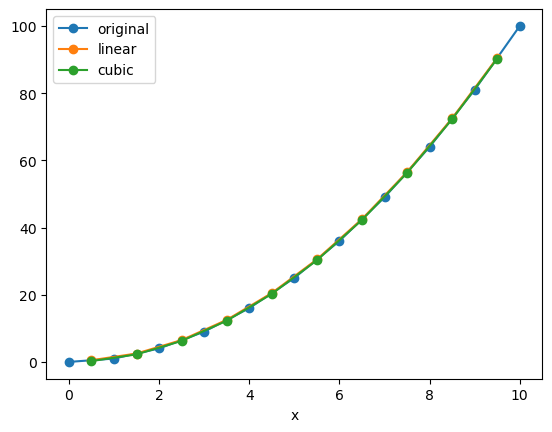
Note that values outside of the original range are not supported:
f_interp_cubic.values
array([ 0.25, 2.25, 6.25, 12.25, 20.25, 30.25, 42.25, 56.25, 72.25,
90.25, nan])
Note
You can apply interpolation to any dimension, and even to multiple dimensions at a time.
(Multidimensional interpolation only supports mode='nearest' and mode='linear'.)
But keep in mind that xarray has no built-in understanding of geography.
If you use interp on lat / lon coordinates, it will just perform naive interpolation of the lat / lon values.
More sophisticated treatment of spherical geometry requires another package such as xesmf.
21.2. Loading Data from NetCDF Files#
NetCDF (Network Common Data Format) is the most widely used format for distributing geoscience data. NetCDF is maintained by the Unidata organization.
Below we quote from the NetCDF website:
NetCDF (network Common Data Form) is a set of interfaces for array-oriented data access and a freely distributed collection of data access libraries for C, Fortran, C++, Java, and other languages. The netCDF libraries support a machine-independent format for representing scientific data. Together, the interfaces, libraries, and format support the creation, access, and sharing of scientific data.
NetCDF data is:
Self-Describing. A netCDF file includes information about the data it contains.
Portable. A netCDF file can be accessed by computers with different ways of storing integers, characters, and floating-point numbers.
Scalable. A small subset of a large dataset may be accessed efficiently.
Appendable. Data may be appended to a properly structured netCDF file without copying the dataset or redefining its structure.
Sharable. One writer and multiple readers may simultaneously access the same netCDF file.
Archivable. Access to all earlier forms of netCDF data will be supported by current and future versions of the software.
xarray is designed to make reading netCDF files in python as easy, powerful, and flexible as possible. (See xarray netCDF docs for more details.)
Below we download and load some the NASA GISSTemp global temperature anomaly dataset. The original file is located at https://data.giss.nasa.gov/pub/gistemp/gistemp1200_GHCNv4_ERSSTv5.nc.gz, but their server is non-responsive at the time of publication of this notebook. So we will use the mirror server from Zenodo.
import pooch
gistemp_file = pooch.retrieve(
'https://zenodo.org/records/13963679/files/gistemp1200_GHCNv4_ERSSTv5.nc',
known_hash=None
)
Downloading data from 'https://zenodo.org/records/13963679/files/gistemp1200_GHCNv4_ERSSTv5.nc' to file '/home/gisuser/.cache/pooch/0e25c9375a619f2b3a461407656b0edc-gistemp1200_GHCNv4_ERSSTv5.nc'.
SHA256 hash of downloaded file: f24422f16fcf86e194308603c989c5a9727d506a6f06125c9abf54b901b004e7
Use this value as the 'known_hash' argument of 'pooch.retrieve' to ensure that the file hasn't changed if it is downloaded again in the future.
ds = xr.open_dataset(gistemp_file)
ds
<xarray.Dataset> Size: 113MB
Dimensions: (lat: 90, lon: 180, time: 1737, nv: 2)
Coordinates:
* lat (lat) float32 360B -89.0 -87.0 -85.0 -83.0 ... 85.0 87.0 89.0
* lon (lon) float32 720B -179.0 -177.0 -175.0 ... 175.0 177.0 179.0
* time (time) datetime64[ns] 14kB 1880-01-15 1880-02-15 ... 2024-09-15
Dimensions without coordinates: nv
Data variables:
time_bnds (time, nv) datetime64[ns] 28kB ...
tempanomaly (time, lat, lon) float32 113MB ...
Attributes:
title: GISTEMP Surface Temperature Analysis
institution: NASA Goddard Institute for Space Studies
source: http://data.giss.nasa.gov/gistemp/
Conventions: CF-1.6
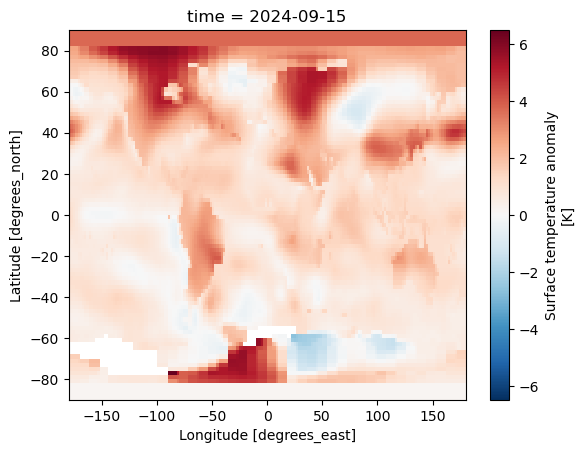
history: Created 2024-10-18 17:43:48 by SBBX_to_nc 2.0 - ILAND=1200,...ds.tempanomaly.isel(time=-1).plot()
plt.show()
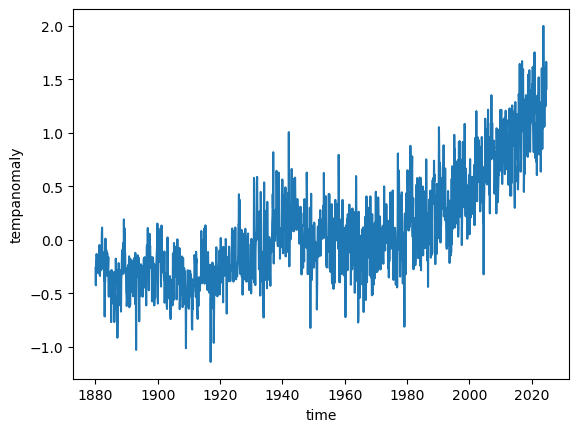
ds.tempanomaly.mean(dim=('lon', 'lat')).plot()
plt.show()
Let’s visualize the temperature anomally for the grid near Worcester. (Worcester’s coordinates: latitude: 42.2626; longtitude: -71.8023)
ds.tempanomaly.sel(lat=42.2626, lon=-71.8023, method='nearest').plot()
plt.show()
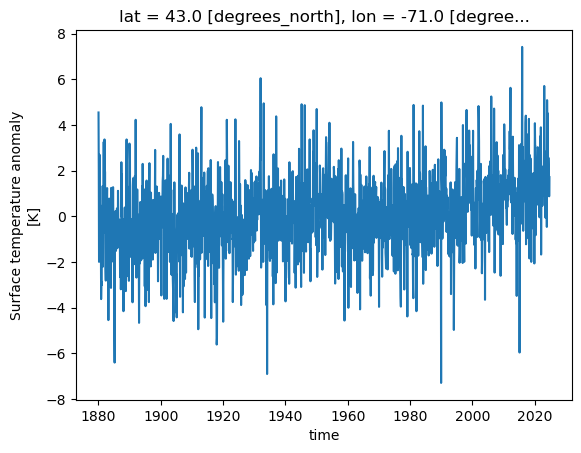
Note that you need to define method since the corresponding values for Worcester latitude and longitude do not exist in the DataArray’s coordinates.
21.3. Groupby#
xarray copies pandas' very useful groupby functionality, enabling the “split / apply / combine” workflow on xarray DataArrays and Datasets. In the first part of the lesson, we will learn to use groupby by analyzing sea-surface temperature data.
First we load a dataset. We will use the NOAA Extended Reconstructed Sea Surface Temperature (ERSST) v5 product, a widely used and trusted gridded compilation of historical data going back to 1854.
Since the data is provided via an OPeNDAP server, we can load it directly without downloading anything:
url = 'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/noaa.ersst.v5/sst.mnmean.nc'
ds = xr.open_dataset(url, drop_variables=['time_bnds'])
ds = ds.sel(time=slice('1960', '2022'))
ds
<xarray.Dataset> Size: 48MB
Dimensions: (lat: 89, lon: 180, time: 756)
Coordinates:
* lat (lat) float32 356B 88.0 86.0 84.0 82.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 720B 0.0 2.0 4.0 6.0 8.0 ... 352.0 354.0 356.0 358.0
* time (time) datetime64[ns] 6kB 1960-01-01 1960-02-01 ... 2022-12-01
Data variables:
sst (time, lat, lon) float32 48MB ...
Attributes: (12/39)
climatology: Climatology is based on 1971-2000 SST, X...
description: In situ data: ICOADS2.5 before 2007 and ...
keywords_vocabulary: NASA Global Change Master Directory (GCM...
keywords: Earth Science > Oceans > Ocean Temperatu...
instrument: Conventional thermometers
source_comment: SSTs were observed by conventional therm...
... ...
comment: SSTs were observed by conventional therm...
summary: ERSST.v5 is developed based on v4 after ...
dataset_title: NOAA Extended Reconstructed SST V5
_NCProperties: version=2,netcdf=4.6.3,hdf5=1.10.5
data_modified: 2024-11-03
DODS_EXTRA.Unlimited_Dimension: timeLet’s do some basic visualizations of the data, just to make sure it looks reasonable.
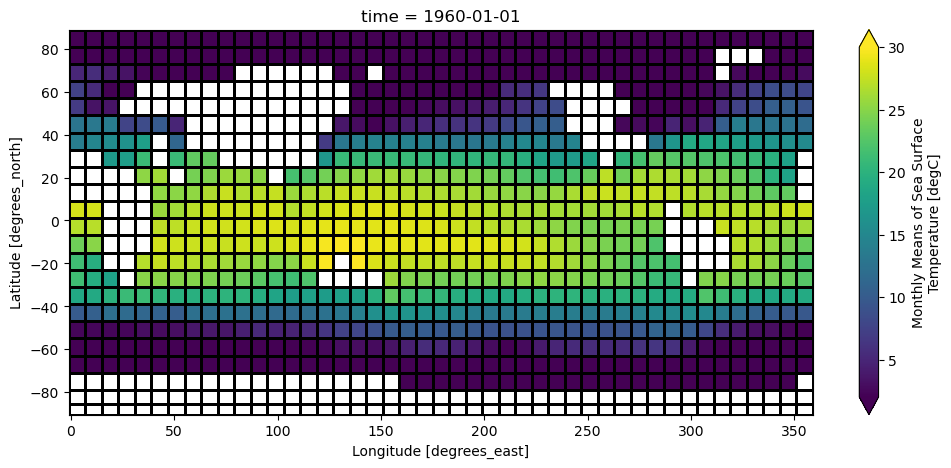
ds.sst.sel(time = '1960-01-01').plot(vmin=-2, vmax=30)
plt.show()
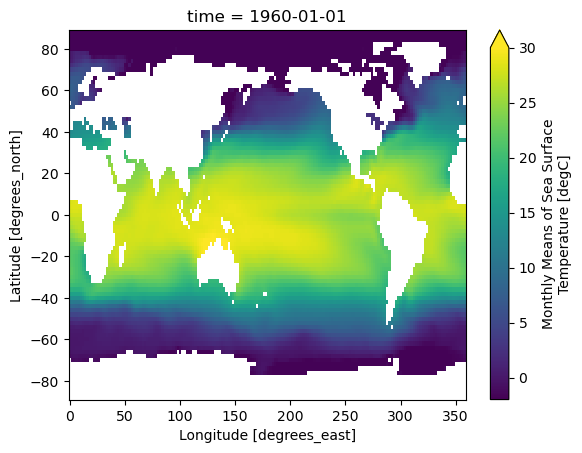
Note that xarray correctly parsed the time index, resulting in a Pandas datetime index on the time dimension.
ds.time
<xarray.DataArray 'time' (time: 756)> Size: 6kB
array(['1960-01-01T00:00:00.000000000', '1960-02-01T00:00:00.000000000',
'1960-03-01T00:00:00.000000000', ..., '2022-10-01T00:00:00.000000000',
'2022-11-01T00:00:00.000000000', '2022-12-01T00:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 6kB 1960-01-01 1960-02-01 ... 2022-12-01
Attributes:
long_name: Time
delta_t: 0000-01-00 00:00:00
avg_period: 0000-01-00 00:00:00
prev_avg_period: 0000-00-07 00:00:00
standard_name: time
axis: T
actual_range: [19723. 82088.]
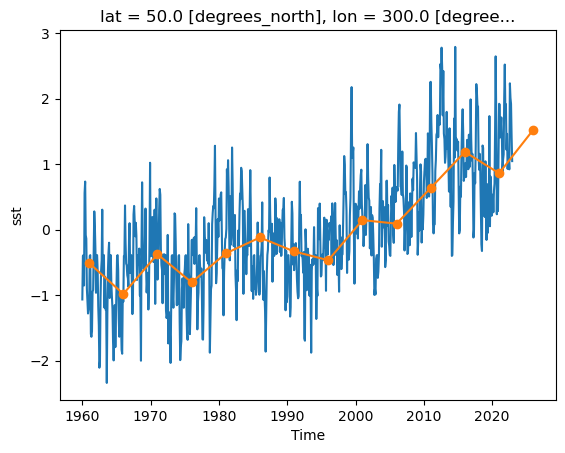
_ChunkSizes: 1ds.sst.sel(lon=300, lat=50).plot()
plt.show()
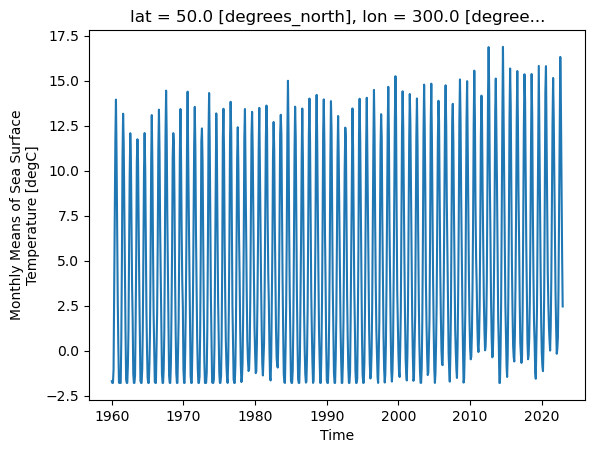
As we can see from the plot, the timeseries at any one point is totally dominated by the seasonal cycle. We would like to remove this seasonal cycle (called the “climatology”) in order to better see the long-term variaitions in temperature. We will accomplish this using groupby.
The syntax of xarray’s groupby is almost identical to pandas.
We will first apply groupby to a single DataArray.
21.3.1. Split Step#
The most important argument is group: this defines the unique values we will use to “split” the data for grouped analysis. We can pass either a DataArray or a name of a variable in the dataset. Lets first use a DataArray. Just like with pandas, we can use the time index to extract specific components of dates and times. xarray uses a special syntax for this .dt, called the DatetimeAccessor.
See a list of datatime properties you can access through .dt here
ds.time.dt
<xarray.core.accessor_dt.DatetimeAccessor at 0xffff46029bd0>
ds.time.dt.month
<xarray.DataArray 'month' (time: 756)> Size: 6kB
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4,
...
6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 11, 12])
Coordinates:
* time (time) datetime64[ns] 6kB 1960-01-01 1960-02-01 ... 2022-12-01
Attributes:
long_name: Time
delta_t: 0000-01-00 00:00:00
avg_period: 0000-01-00 00:00:00
prev_avg_period: 0000-00-07 00:00:00
standard_name: time
axis: T
actual_range: [19723. 82088.]
_ChunkSizes: 1ds.time.dt.year
<xarray.DataArray 'year' (time: 756)> Size: 6kB
array([1960, 1960, 1960, 1960, 1960, 1960, 1960, 1960, 1960, 1960, 1960,
1960, 1961, 1961, 1961, 1961, 1961, 1961, 1961, 1961, 1961, 1961,
1961, 1961, 1962, 1962, 1962, 1962, 1962, 1962, 1962, 1962, 1962,
1962, 1962, 1962, 1963, 1963, 1963, 1963, 1963, 1963, 1963, 1963,
1963, 1963, 1963, 1963, 1964, 1964, 1964, 1964, 1964, 1964, 1964,
1964, 1964, 1964, 1964, 1964, 1965, 1965, 1965, 1965, 1965, 1965,
1965, 1965, 1965, 1965, 1965, 1965, 1966, 1966, 1966, 1966, 1966,
1966, 1966, 1966, 1966, 1966, 1966, 1966, 1967, 1967, 1967, 1967,
1967, 1967, 1967, 1967, 1967, 1967, 1967, 1967, 1968, 1968, 1968,
1968, 1968, 1968, 1968, 1968, 1968, 1968, 1968, 1968, 1969, 1969,
1969, 1969, 1969, 1969, 1969, 1969, 1969, 1969, 1969, 1969, 1970,
1970, 1970, 1970, 1970, 1970, 1970, 1970, 1970, 1970, 1970, 1970,
1971, 1971, 1971, 1971, 1971, 1971, 1971, 1971, 1971, 1971, 1971,
1971, 1972, 1972, 1972, 1972, 1972, 1972, 1972, 1972, 1972, 1972,
1972, 1972, 1973, 1973, 1973, 1973, 1973, 1973, 1973, 1973, 1973,
1973, 1973, 1973, 1974, 1974, 1974, 1974, 1974, 1974, 1974, 1974,
1974, 1974, 1974, 1974, 1975, 1975, 1975, 1975, 1975, 1975, 1975,
1975, 1975, 1975, 1975, 1975, 1976, 1976, 1976, 1976, 1976, 1976,
1976, 1976, 1976, 1976, 1976, 1976, 1977, 1977, 1977, 1977, 1977,
1977, 1977, 1977, 1977, 1977, 1977, 1977, 1978, 1978, 1978, 1978,
...
2004, 2005, 2005, 2005, 2005, 2005, 2005, 2005, 2005, 2005, 2005,
2005, 2005, 2006, 2006, 2006, 2006, 2006, 2006, 2006, 2006, 2006,
2006, 2006, 2006, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007,
2007, 2007, 2007, 2007, 2008, 2008, 2008, 2008, 2008, 2008, 2008,
2008, 2008, 2008, 2008, 2008, 2009, 2009, 2009, 2009, 2009, 2009,
2009, 2009, 2009, 2009, 2009, 2009, 2010, 2010, 2010, 2010, 2010,
2010, 2010, 2010, 2010, 2010, 2010, 2010, 2011, 2011, 2011, 2011,
2011, 2011, 2011, 2011, 2011, 2011, 2011, 2011, 2012, 2012, 2012,
2012, 2012, 2012, 2012, 2012, 2012, 2012, 2012, 2012, 2013, 2013,
2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2014,
2014, 2014, 2014, 2014, 2014, 2014, 2014, 2014, 2014, 2014, 2014,
2015, 2015, 2015, 2015, 2015, 2015, 2015, 2015, 2015, 2015, 2015,
2015, 2016, 2016, 2016, 2016, 2016, 2016, 2016, 2016, 2016, 2016,
2016, 2016, 2017, 2017, 2017, 2017, 2017, 2017, 2017, 2017, 2017,
2017, 2017, 2017, 2018, 2018, 2018, 2018, 2018, 2018, 2018, 2018,
2018, 2018, 2018, 2018, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2020, 2020, 2020, 2020, 2020, 2020,
2020, 2020, 2020, 2020, 2020, 2020, 2021, 2021, 2021, 2021, 2021,
2021, 2021, 2021, 2021, 2021, 2021, 2021, 2022, 2022, 2022, 2022,
2022, 2022, 2022, 2022, 2022, 2022, 2022, 2022])
Coordinates:
* time (time) datetime64[ns] 6kB 1960-01-01 1960-02-01 ... 2022-12-01
Attributes:
long_name: Time
delta_t: 0000-01-00 00:00:00
avg_period: 0000-01-00 00:00:00
prev_avg_period: 0000-00-07 00:00:00
standard_name: time
axis: T
actual_range: [19723. 82088.]
_ChunkSizes: 1We can use these arrays in a groupby operation:
gb = ds.sst.groupby(ds.time.dt.month)
gb
<DataArrayGroupBy, grouped over 1 grouper(s), 12 groups in total:
'month': 12/12 groups present with labels 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12>
xarray also offers a more concise syntax when the variable you’re grouping on is already present in the dataset. This is identical to the previous line:
gb = ds.sst.groupby('time.month')
gb
<DataArrayGroupBy, grouped over 1 grouper(s), 12 groups in total:
'month': 12/12 groups present with labels 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12>
Now that the data are split, we can manually iterate over the group. The iterator returns the key (group name) and the value (the actual dataset corresponding to that group) for each group.
for group_name, group_da in gb:
print(group_name) # stop after first iteration
break
group_da
1
<xarray.DataArray 'sst' (time: 63, lat: 89, lon: 180)> Size: 4MB
[1009260 values with dtype=float32]
Coordinates:
* lat (lat) float32 356B 88.0 86.0 84.0 82.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 720B 0.0 2.0 4.0 6.0 8.0 ... 352.0 354.0 356.0 358.0
* time (time) datetime64[ns] 504B 1960-01-01 1961-01-01 ... 2022-01-01
Attributes:
long_name: Monthly Means of Sea Surface Temperature
units: degC
var_desc: Sea Surface Temperature
level_desc: Surface
statistic: Mean
dataset: NOAA Extended Reconstructed SST V5
parent_stat: Individual Values
actual_range: [-1.8 42.32636]
valid_range: [-1.8 45. ]
_ChunkSizes: [ 1 89 180]21.3.2. Map & Combine#
Now that we have groups defined, it’s time to “apply” a calculation to the group. Like in pandas, these calculations can either be:
aggregation: reduces the size of the group
transformation: preserves the group’s full size
At then end of the apply step, xarray will automatically combine the aggregated / transformed groups back into a single object.
Warning
Xarray calls the “apply” step map. This is different from Pandas!
gb.map?
Signature:
gb.map(
func: 'Callable[..., DataArray]',
args: 'tuple[Any, ...]' = (),
shortcut: 'bool | None' = None,
**kwargs: 'Any',
) -> 'DataArray'
Docstring:
Apply a function to each array in the group and concatenate them
together into a new array.
`func` is called like `func(ar, *args, **kwargs)` for each array `ar`
in this group.
Apply uses heuristics (like `pandas.GroupBy.apply`) to figure out how
to stack together the array. The rule is:
1. If the dimension along which the group coordinate is defined is
still in the first grouped array after applying `func`, then stack
over this dimension.
2. Otherwise, stack over the new dimension given by name of this
grouping (the argument to the `groupby` function).
Parameters
----------
func : callable
Callable to apply to each array.
shortcut : bool, optional
Whether or not to shortcut evaluation under the assumptions that:
(1) The action of `func` does not depend on any of the array
metadata (attributes or coordinates) but only on the data and
dimensions.
(2) The action of `func` creates arrays with homogeneous metadata,
that is, with the same dimensions and attributes.
If these conditions are satisfied `shortcut` provides significant
speedup. This should be the case for many common groupby operations
(e.g., applying numpy ufuncs).
*args : tuple, optional
Positional arguments passed to `func`.
**kwargs
Used to call `func(ar, **kwargs)` for each array `ar`.
Returns
-------
applied : DataArray
The result of splitting, applying and combining this array.
File: /opt/conda/envs/xarray_tutorial/lib/python3.13/site-packages/xarray/core/groupby.py
Type: method
21.3.2.1. Aggregations#
Like pandas, xarray’s groupby object has many built-in aggregation operations (e.g. mean, min, max, std, etc):
sst_mm = gb.mean(dim='time')
sst_mm
<xarray.DataArray 'sst' (month: 12, lat: 89, lon: 180)> Size: 769kB
array([[[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
...
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[-1.7995719, -1.7996585, -1.7998686, ..., -1.7998053,
-1.7996899, -1.7995679],
[-1.799625 , -1.7997937, -1.800001 , ..., -1.800001 ,
-1.7998347, -1.7996482],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]]], dtype=float32)
Coordinates:
* lat (lat) float32 356B 88.0 86.0 84.0 82.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 720B 0.0 2.0 4.0 6.0 8.0 ... 352.0 354.0 356.0 358.0
* month (month) int64 96B 1 2 3 4 5 6 7 8 9 10 11 12
Attributes:
long_name: Monthly Means of Sea Surface Temperature
units: degC
var_desc: Sea Surface Temperature
level_desc: Surface
statistic: Mean
dataset: NOAA Extended Reconstructed SST V5
parent_stat: Individual Values
actual_range: [-1.8 42.32636]
valid_range: [-1.8 45. ]
_ChunkSizes: [ 1 89 180].map accepts as its argument a function. We can pass an existing function:
gb.map(np.mean)
<xarray.DataArray 'sst' (month: 12)> Size: 48B
array([13.679474, 13.787702, 13.784417, 13.704169, 13.662362, 13.736686,
13.950283, 14.123172, 14.008736, 13.715602, 13.528554, 13.548446],
dtype=float32)
Coordinates:
* month (month) int64 96B 1 2 3 4 5 6 7 8 9 10 11 12Because we specified no extra arguments (like axis) the function was applied over all space and time dimensions. This is not what we wanted. Instead, we could define a custom function. This function takes a single argument –the group dataset– and returns a new dataset to be combined:
def time_mean(a):
return a.mean(dim='time')
sst_mm = gb.map(time_mean)
sst_mm
<xarray.DataArray 'sst' (month: 12, lat: 89, lon: 180)> Size: 769kB
array([[[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
...
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[-1.7995719, -1.7996585, -1.7998686, ..., -1.7998053,
-1.7996899, -1.7995679],
[-1.799625 , -1.7997937, -1.800001 , ..., -1.800001 ,
-1.7998347, -1.7996482],
[-1.800001 , -1.800001 , -1.800001 , ..., -1.800001 ,
-1.800001 , -1.800001 ],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]]], dtype=float32)
Coordinates:
* lat (lat) float32 356B 88.0 86.0 84.0 82.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 720B 0.0 2.0 4.0 6.0 8.0 ... 352.0 354.0 356.0 358.0
* month (month) int64 96B 1 2 3 4 5 6 7 8 9 10 11 12So we did what we wanted to do: calculate the climatology at every point in the dataset. Let’s look at the data a bit.
Climatlogy at a specific point in the North Atlantic
sst_mm.sel(lon=300, lat=-50).plot()
plt.show()
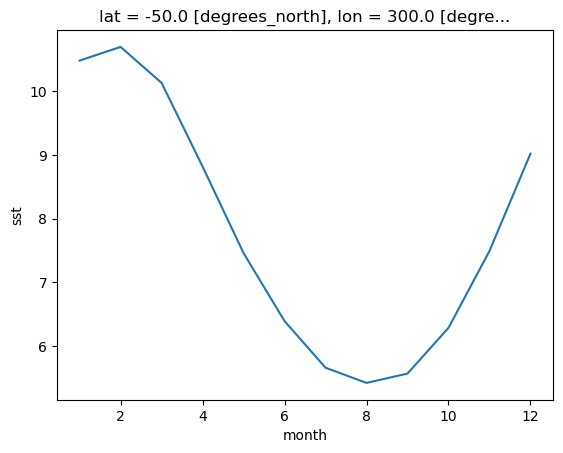
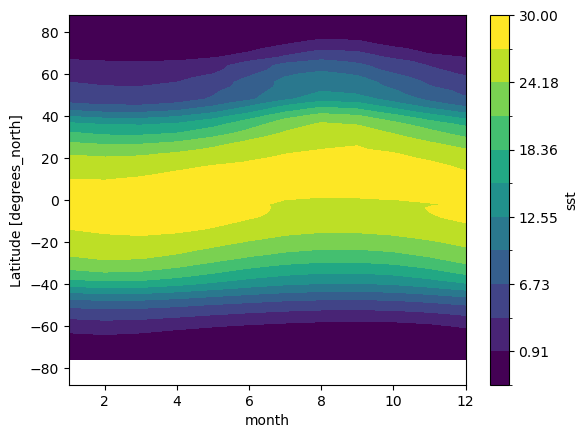
Zonal Mean Climatology
sst_mm.mean(dim='lon').transpose().plot.contourf(levels=12, vmin=-2, vmax=30)
plt.show()
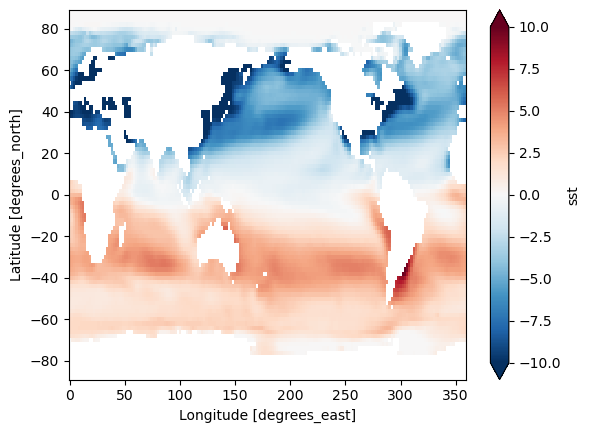
Difference between January and July Climatology
(sst_mm.sel(month=1) - sst_mm.sel(month=7)).plot(vmax=10)
plt.show()
21.3.2.2. Transformations#
Now we want to remove this climatology from the dataset, to examine the residual, called the anomaly, which is the interesting part from a climate perspective. Removing the seasonal climatology is a perfect example of a transformation: it operates over a group, but doesn’t change the size of the dataset. Here is one way to code it.
def remove_time_mean(x):
return x - x.mean(dim='time')
ds_anom = ds.groupby('time.month').map(remove_time_mean)
ds_anom
<xarray.Dataset> Size: 48MB
Dimensions: (time: 756, lat: 89, lon: 180)
Coordinates:
* lat (lat) float32 356B 88.0 86.0 84.0 82.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 720B 0.0 2.0 4.0 6.0 8.0 ... 352.0 354.0 356.0 358.0
* time (time) datetime64[ns] 6kB 1960-01-01 1960-02-01 ... 2022-12-01
Data variables:
sst (time, lat, lon) float32 48MB 1.073e-06 1.073e-06 ... nan nanNote
In the above example, we applied groupby to a Dataset instead of a DataArray.
Xarray makes these sorts of transformations easy by supporting groupby arithmetic. This concept is easiest explained with an example:
gb = ds.groupby('time.month')
ds_anom = gb - gb.mean(dim='time')
ds_anom
<xarray.Dataset> Size: 48MB
Dimensions: (lat: 89, lon: 180, time: 756)
Coordinates:
* lat (lat) float32 356B 88.0 86.0 84.0 82.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 720B 0.0 2.0 4.0 6.0 8.0 ... 352.0 354.0 356.0 358.0
* time (time) datetime64[ns] 6kB 1960-01-01 1960-02-01 ... 2022-12-01
month (time) int64 6kB 1 2 3 4 5 6 7 8 9 10 11 ... 3 4 5 6 7 8 9 10 11 12
Data variables:
sst (time, lat, lon) float32 48MB 1.073e-06 1.073e-06 ... nan nanNow we can view the climate signal without the overwhelming influence of the seasonal cycle.
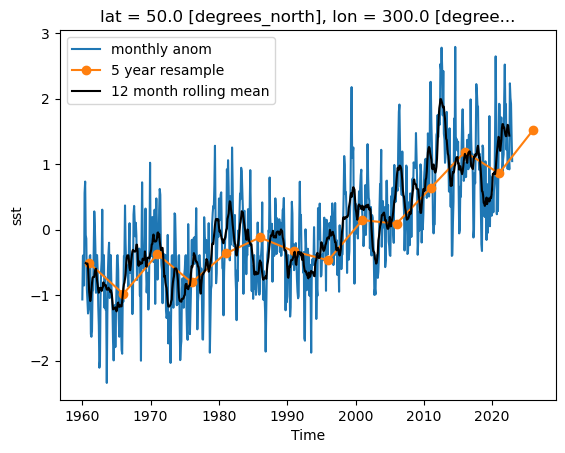
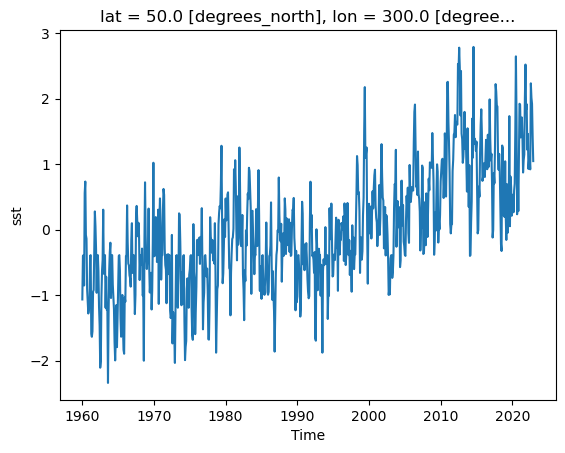
Timeseries at a single point in the North Atlantic
ds_anom.sst.sel(lon=300, lat=50).plot()
plt.show()
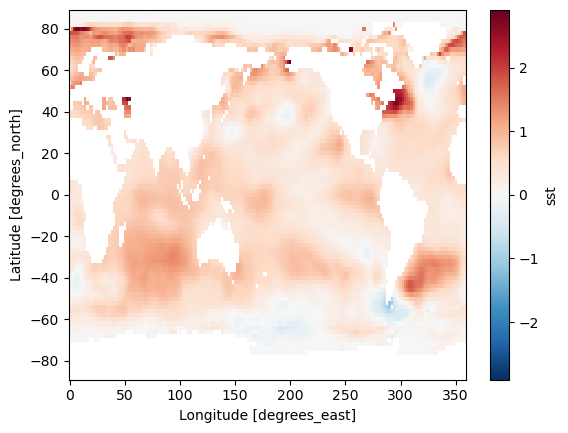
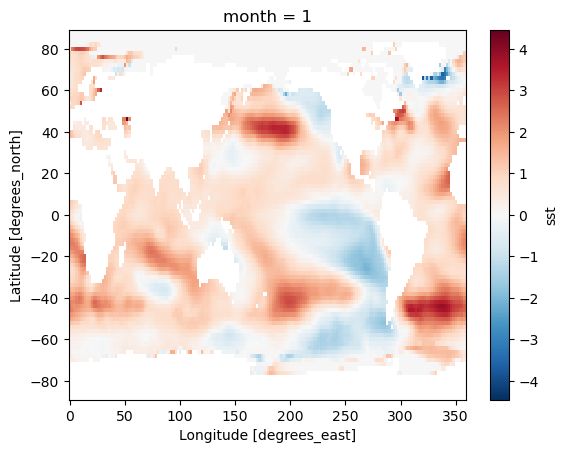
Difference between Jan. 1 2022 and Jan. 1 1960
(ds_anom.sel(time='2022-01-01') - ds_anom.sel(time='1960-01-01')).sst.plot()
plt.show()
21.5. Coarsen#
Coarsen is a simple way to reduce the size of your data along one or more axes.
It is very similar to resample when operating on time dimensions; the key difference is that coarsen only operates on fixed blocks of data, irrespective of the coordinate values, while resample actually looks at the coordinates to figure out, e.g. what month a particular data point is in.
For regularly-spaced monthly data beginning in January, the following should be equivalent to annual resampling. However, results would be different for irregularly-spaced data.
ds.coarsen(time=12, boundary = 'exact').mean()
<xarray.Dataset> Size: 4MB
Dimensions: (time: 63, lat: 89, lon: 180)
Coordinates:
* lat (lat) float32 356B 88.0 86.0 84.0 82.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 720B 0.0 2.0 4.0 6.0 8.0 ... 352.0 354.0 356.0 358.0
* time (time) datetime64[ns] 504B 1960-06-16T08:00:00 ... 2022-06-16T12...
Data variables:
sst (time, lat, lon) float32 4MB -1.8 -1.8 -1.8 -1.8 ... nan nan nan
Attributes: (12/39)
climatology: Climatology is based on 1971-2000 SST, X...
description: In situ data: ICOADS2.5 before 2007 and ...
keywords_vocabulary: NASA Global Change Master Directory (GCM...
keywords: Earth Science > Oceans > Ocean Temperatu...
instrument: Conventional thermometers
source_comment: SSTs were observed by conventional therm...
... ...
comment: SSTs were observed by conventional therm...
summary: ERSST.v5 is developed based on v4 after ...
dataset_title: NOAA Extended Reconstructed SST V5
_NCProperties: version=2,netcdf=4.6.3,hdf5=1.10.5
data_modified: 2024-11-03
DODS_EXTRA.Unlimited_Dimension: timeCoarsen also works on spatial coordinates (or any coordiantes).
ds_coarse = ds.coarsen(lon=4, lat=4, boundary='pad').mean()
ds_coarse.sst.isel(time=0).plot(vmin=2, vmax=30, figsize=(12, 5), edgecolor='k')
plt.show()