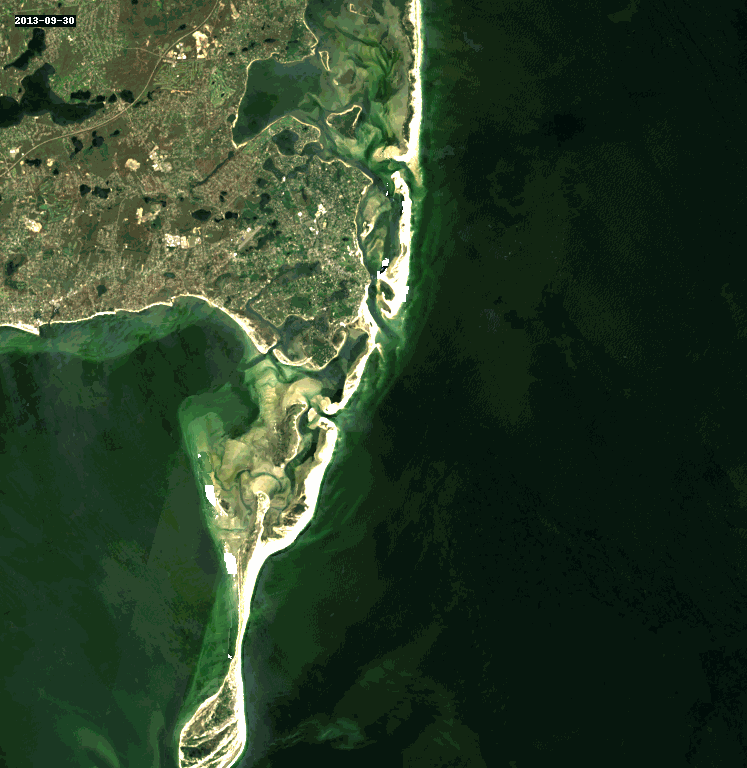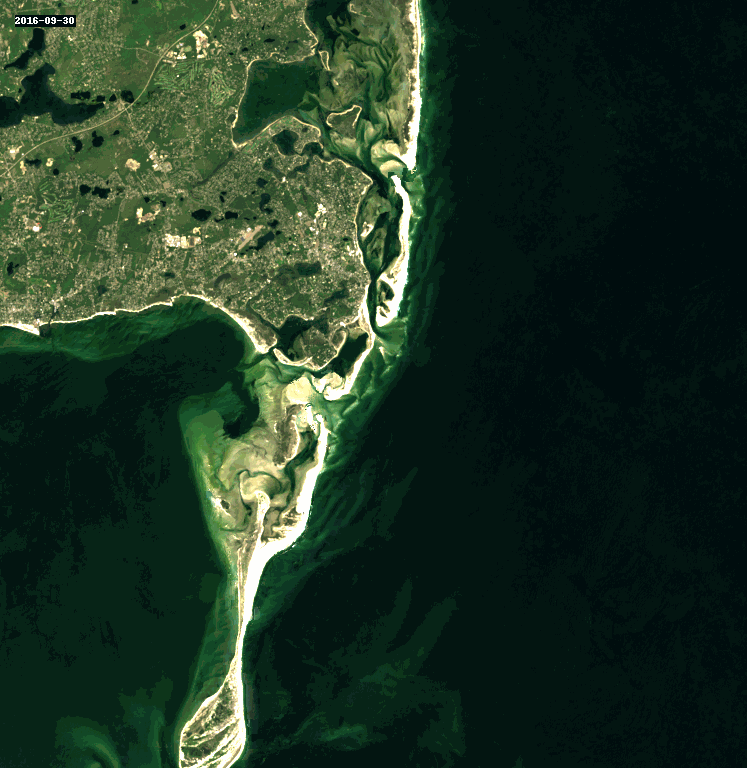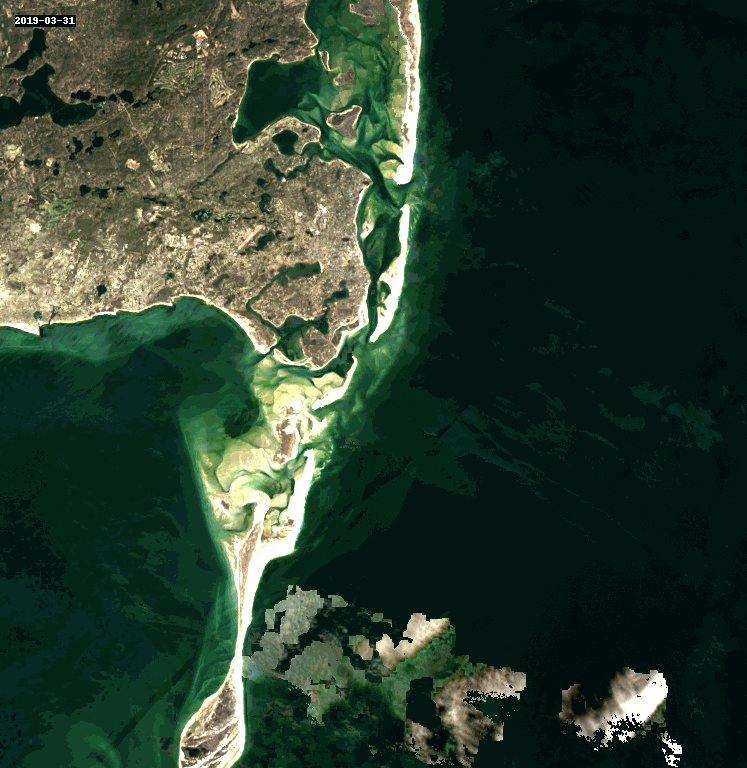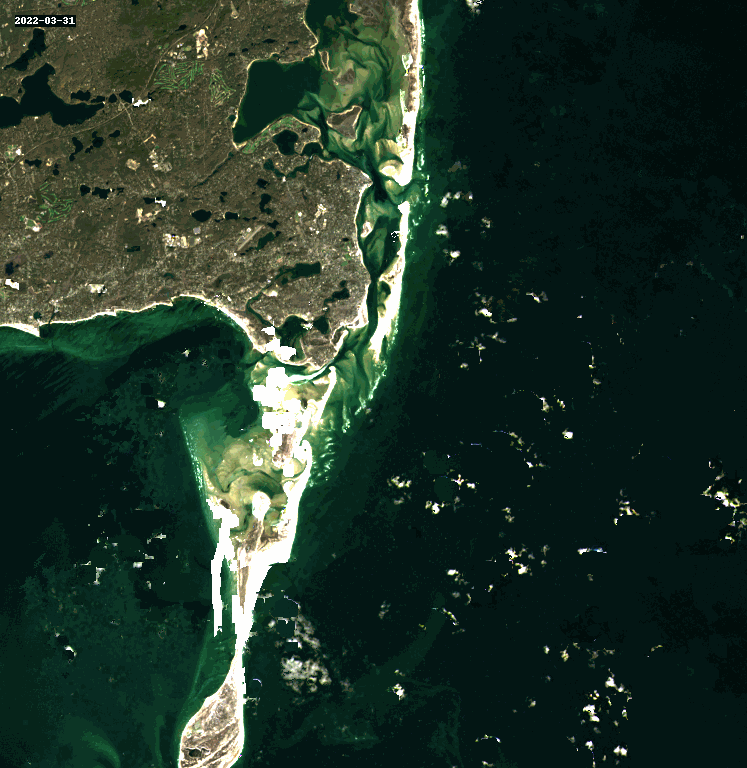
11. Query and Stack Imagery into a GIF Using stackstac and Dask#
Attribution: This tutorial is developed based on the example provided in the stackstac documentation.
We’ll load all the Landsat-8 (Collection 2, Level 2) data that’s available from Microsoft’s Planetary Computer over a small region on the coast of Cape Cod, Massachusetts, USA.
Using nothing but standard xarray syntax, we’ll mask cloudy pixels with the Landsat QA band and reduce the data down to biannual median composites. We will use Dask to make this much faster.
Animated as a GIF, we can watch the coastline move over the years due to longshore drift.
You can access this notebook (in a Docker image) on this GitHub repo.
import dask
import pystac_client
import planetary_computer
import leafmap
import geogif
import stackstac
Start your Dask cluster:
from dask.distributed import Client, LocalCluster
cluster = LocalCluster()
client = Client(cluster)
client
Client
Client-6607a032-8729-11ef-80fe-0242ac110002
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:8787/status |
Cluster Info
LocalCluster
f5532b7f
| Dashboard: http://127.0.0.1:8787/status | Workers: 5 |
| Total threads: 5 | Total memory: 7.67 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-00953ed9-9f76-4f5c-8a21-f27ca5f46b48
| Comm: tcp://127.0.0.1:40851 | Workers: 5 |
| Dashboard: http://127.0.0.1:8787/status | Total threads: 5 |
| Started: Just now | Total memory: 7.67 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:45323 | Total threads: 1 |
| Dashboard: http://127.0.0.1:39459/status | Memory: 1.53 GiB |
| Nanny: tcp://127.0.0.1:33483 | |
| Local directory: /tmp/dask-worker-space/worker-0sm5aun5 | |
Worker: 1
| Comm: tcp://127.0.0.1:39923 | Total threads: 1 |
| Dashboard: http://127.0.0.1:42603/status | Memory: 1.53 GiB |
| Nanny: tcp://127.0.0.1:35887 | |
| Local directory: /tmp/dask-worker-space/worker-m5hkut_g | |
Worker: 2
| Comm: tcp://127.0.0.1:35045 | Total threads: 1 |
| Dashboard: http://127.0.0.1:42285/status | Memory: 1.53 GiB |
| Nanny: tcp://127.0.0.1:35307 | |
| Local directory: /tmp/dask-worker-space/worker-uk_b1erf | |
Worker: 3
| Comm: tcp://127.0.0.1:38185 | Total threads: 1 |
| Dashboard: http://127.0.0.1:42747/status | Memory: 1.53 GiB |
| Nanny: tcp://127.0.0.1:46121 | |
| Local directory: /tmp/dask-worker-space/worker-1j__7a1l | |
Worker: 4
| Comm: tcp://127.0.0.1:46875 | Total threads: 1 |
| Dashboard: http://127.0.0.1:43369/status | Memory: 1.53 GiB |
| Nanny: tcp://127.0.0.1:46023 | |
| Local directory: /tmp/dask-worker-space/worker-pm6377o2 | |
Interactively pick the area of interest from a map. Just move the map around and re-run all cells to generate the timeseries somewhere else!
import leafmap
m = leafmap.Map(center=[41.64933994767867, -69.94438630063088], zoom=12, height="800px")
m
bbox = (m.west, m.south, m.east, m.north)
11.1. Search for STAC items#
Use pystac-client to connect to Microsoft’s STAC API endpoint and search for Landsat-8 scenes.
catalog = pystac_client.Client.open(
"https://planetarycomputer.microsoft.com/api/stac/v1",
modifier=planetary_computer.sign_inplace,
)
search = catalog.search(
collections=['landsat-8-c2-l2'],
bbox=bbox,
)
Load all the STAC items:
items = search.get_all_items()
len(items)
201
These are the footprints of all the items we’ll use:
m.add_geojson(items.to_dict(), zoom=12)
m
11.2. Create an xarray with stacksatc#
Set bounds_latlon=bbox to automatically clip to our area of interest (instead of using the full footprints of the scenes).
%%time
stack = stackstac.stack(items, bounds_latlon=bbox)
stack
CPU times: user 104 ms, sys: 4.8 ms, total: 109 ms
Wall time: 106 ms
<xarray.DataArray 'stackstac-0bffcf6778474275db65519a313c614c' (time: 201,
band: 19,
y: 768, x: 747)>
dask.array<fetch_raster_window, shape=(201, 19, 768, 747), dtype=float64, chunksize=(1, 1, 768, 747), chunktype=numpy.ndarray>
Coordinates: (12/27)
* time (time) datetime64[ns] 2013-03-22T15:19:00.54...
id (time) <U31 'LC08_L2SP_011031_20130322_02_T1...
* band (band) <U13 'SR_B1' 'SR_B2' ... 'SR_QA_AEROSOL'
* x (x) float64 4.101e+05 4.102e+05 ... 4.325e+05
* y (y) float64 4.623e+06 4.623e+06 ... 4.6e+06
view:off_nadir int64 0
... ...
title (band) <U46 'Coastal/Aerosol Band (B1)' ... ...
gsd (band) float64 30.0 30.0 30.0 ... 30.0 30.0
common_name (band) object 'coastal' 'blue' ... None None
center_wavelength (band) object 0.44 0.48 0.56 ... None None None
full_width_half_max (band) object 0.02 0.06 0.06 ... None None None
epsg int64 32619
Attributes:
spec: RasterSpec(epsg=32619, bounds=(410130.0, 4599750.0, 432540.0...
crs: epsg:32619
transform: | 30.00, 0.00, 410130.00|\n| 0.00,-30.00, 4622790.00|\n| 0.0...
resolution: 30.0And that’s it for stackstac! Everything from here on is just standard xarray operations.
Band names are not trivial from the initial query:
stack.band
<xarray.DataArray 'band' (band: 19)>
array(['SR_B1', 'SR_B2', 'SR_B3', 'SR_B4', 'SR_B5', 'SR_B6', 'SR_B7', 'ST_QA',
'ST_B10', 'ST_DRAD', 'ST_EMIS', 'ST_EMSD', 'ST_TRAD', 'ST_URAD',
'QA_PIXEL', 'ST_ATRAN', 'ST_CDIST', 'QA_RADSAT', 'SR_QA_AEROSOL'],
dtype='<U13')
Coordinates: (12/16)
* band (band) <U13 'SR_B1' 'SR_B2' ... 'SR_QA_AEROSOL'
view:off_nadir int64 0
instruments object {'oli', 'tirs'}
landsat:collection_number <U2 '02'
platform <U9 'landsat-8'
description (band) <U91 'Collection 2 Level-2 Coastal/Aero...
... ...
title (band) <U46 'Coastal/Aerosol Band (B1)' ... 'A...
gsd (band) float64 30.0 30.0 30.0 ... 30.0 30.0 30.0
common_name (band) object 'coastal' 'blue' ... None None
center_wavelength (band) object 0.44 0.48 0.56 ... None None None
full_width_half_max (band) object 0.02 0.06 0.06 ... None None None
epsg int64 32619You can replace them using the common_name field:
stack = stack.assign_coords(band=stack.common_name.fillna(stack.band).rename("band"))
stack.band
<xarray.DataArray 'band' (band: 19)>
array(['coastal', 'blue', 'green', 'red', 'nir08', 'swir16', 'swir22', 'ST_QA',
'lwir11', 'ST_DRAD', 'ST_EMIS', 'ST_EMSD', 'ST_TRAD', 'ST_URAD',
'QA_PIXEL', 'ST_ATRAN', 'ST_CDIST', 'QA_RADSAT', 'SR_QA_AEROSOL'],
dtype=object)
Coordinates: (12/16)
view:off_nadir int64 0
instruments object {'oli', 'tirs'}
landsat:collection_number <U2 '02'
platform <U9 'landsat-8'
description (band) <U91 'Collection 2 Level-2 Coastal/Aero...
landsat:processing_level <U4 'L2SP'
... ...
gsd (band) float64 30.0 30.0 30.0 ... 30.0 30.0 30.0
common_name (band) object 'coastal' 'blue' ... None None
center_wavelength (band) object 0.44 0.48 0.56 ... None None None
full_width_half_max (band) object 0.02 0.06 0.06 ... None None None
epsg int64 32619
* band (band) object 'coastal' ... 'SR_QA_AEROSOL'See how much input data there is for just RGB. This is the amount of data we’ll end up processing
stack.sel(band=["red", "green", "blue"])
<xarray.DataArray 'stackstac-0bffcf6778474275db65519a313c614c' (time: 201,
band: 3,
y: 768, x: 747)>
dask.array<getitem, shape=(201, 3, 768, 747), dtype=float64, chunksize=(1, 1, 768, 747), chunktype=numpy.ndarray>
Coordinates: (12/27)
* time (time) datetime64[ns] 2013-03-22T15:19:00.54...
id (time) <U31 'LC08_L2SP_011031_20130322_02_T1...
* x (x) float64 4.101e+05 4.102e+05 ... 4.325e+05
* y (y) float64 4.623e+06 4.623e+06 ... 4.6e+06
view:off_nadir int64 0
instruments object {'oli', 'tirs'}
... ...
gsd (band) float64 30.0 30.0 30.0
common_name (band) object 'red' 'green' 'blue'
center_wavelength (band) object 0.65 0.56 0.48
full_width_half_max (band) object 0.04 0.06 0.06
epsg int64 32619
* band (band) object 'red' 'green' 'blue'
Attributes:
spec: RasterSpec(epsg=32619, bounds=(410130.0, 4599750.0, 432540.0...
crs: epsg:32619
transform: | 30.00, 0.00, 410130.00|\n| 0.00,-30.00, 4622790.00|\n| 0.0...
resolution: 30.011.3. Mask cloudy pixels using the QA band#
Use the bit values of the Landsat-8 QA band to mask out bad pixels. We’ll mask pixels labeled as dilated cloud, cirrus, cloud, or cloud shadow. (By “mask”, we mean just replacing those pixels with NaNs).
See page 14 on this PDF for the data table describing which bit means what.
mask_bitfields = [1, 2, 3, 4] # dilated cloud, cirrus, cloud, cloud shadow
bitmask = 0
for field in mask_bitfields:
bitmask |= 1 << field
bin(bitmask)
'0b11110'
qa = stack.sel(band="QA_PIXEL").astype("uint16")
mask = qa & bitmask # just look at those 4 bits
cloud_free_stack = stack.where(mask == 0) # mask pixels where any one of those bits are set
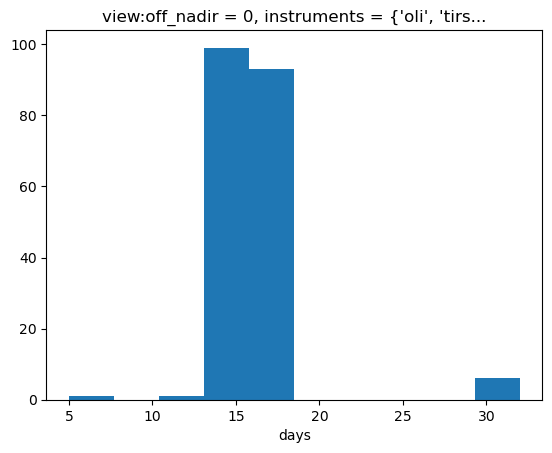
# What's the typical interval between scenes?
cloud_free_stack.time.diff("time").dt.days.plot.hist();
11.4. Make biannual median composites#
The Landsat-8 scenes appear to typically be 5-15 days apart. Let’s composite that down to a 6-month interval.
Since the cloudy pixels we masked with NaNs will be ignored in the median, this should give us a decent cloud-free-ish image for each.
# Make biannual median composites (`2Q` means 2 quarters)
composites_stack = cloud_free_stack.resample(time="2Q").median("time")
composites_stack
<xarray.DataArray 'stackstac-0bffcf6778474275db65519a313c614c' (time: 19,
band: 19,
y: 768, x: 747)>
dask.array<stack, shape=(19, 19, 768, 747), dtype=float64, chunksize=(1, 1, 768, 747), chunktype=numpy.ndarray>
Coordinates: (12/13)
* x (x) float64 4.101e+05 4.102e+05 ... 4.325e+05
* y (y) float64 4.623e+06 4.623e+06 ... 4.6e+06
view:off_nadir int64 0
instruments object {'oli', 'tirs'}
landsat:collection_number <U2 '02'
platform <U9 'landsat-8'
... ...
proj:epsg int64 32619
landsat:wrs_type <U1 '2'
landsat:wrs_row <U3 '031'
epsg int64 32619
* band (band) object 'coastal' ... 'SR_QA_AEROSOL'
* time (time) datetime64[ns] 2013-03-31 ... 2022-03-31
Attributes:
spec: RasterSpec(epsg=32619, bounds=(410130.0, 4599750.0, 432540.0...
crs: epsg:32619
transform: | 30.00, 0.00, 410130.00|\n| 0.00,-30.00, 4622790.00|\n| 0.0...
resolution: 30.0Pick the red-green-blue bands to make a true-color image.
rgb_stack = composites_stack.sel(band=["red", "green", "blue"])
rgb_stack
<xarray.DataArray 'stackstac-0bffcf6778474275db65519a313c614c' (time: 19,
band: 3,
y: 768, x: 747)>
dask.array<getitem, shape=(19, 3, 768, 747), dtype=float64, chunksize=(1, 1, 768, 747), chunktype=numpy.ndarray>
Coordinates: (12/13)
* x (x) float64 4.101e+05 4.102e+05 ... 4.325e+05
* y (y) float64 4.623e+06 4.623e+06 ... 4.6e+06
view:off_nadir int64 0
instruments object {'oli', 'tirs'}
landsat:collection_number <U2 '02'
platform <U9 'landsat-8'
... ...
proj:epsg int64 32619
landsat:wrs_type <U1 '2'
landsat:wrs_row <U3 '031'
epsg int64 32619
* band (band) object 'red' 'green' 'blue'
* time (time) datetime64[ns] 2013-03-31 ... 2022-03-31
Attributes:
spec: RasterSpec(epsg=32619, bounds=(410130.0, 4599750.0, 432540.0...
crs: epsg:32619
transform: | 30.00, 0.00, 410130.00|\n| 0.00,-30.00, 4622790.00|\n| 0.0...
resolution: 30.0Some final cleanup to make a nicer-looking animation:
Forward-fill any NaN pixels from the previous frame, to make the animation look less jumpy.
Also skip the first frame, since its NaNs can’t be filled from anywhere.
cleaned_stack = rgb_stack.ffill("time")[1:]
11.5. Render the GIF#
Use GeoGIF to turn the stack into an animation. We’ll use dgif to render the GIF on the cluster, so there’s less data to send back. (GIFs are a lot smaller than NumPy arrays!)
%%time
gif_img = geogif.dgif(cleaned_stack).compute()
/opt/conda/lib/python3.10/site-packages/numpy/lib/nanfunctions.py:1217: RuntimeWarning: All-NaN slice encountered
r, k = function_base._ureduce(a, func=_nanmedian, axis=axis, out=out,
/opt/conda/lib/python3.10/site-packages/numpy/lib/nanfunctions.py:1217: RuntimeWarning: All-NaN slice encountered
r, k = function_base._ureduce(a, func=_nanmedian, axis=axis, out=out,
/opt/conda/lib/python3.10/site-packages/numpy/lib/nanfunctions.py:1217: RuntimeWarning: All-NaN slice encountered
r, k = function_base._ureduce(a, func=_nanmedian, axis=axis, out=out,
/opt/conda/lib/python3.10/site-packages/numpy/lib/nanfunctions.py:1217: RuntimeWarning: All-NaN slice encountered
r, k = function_base._ureduce(a, func=_nanmedian, axis=axis, out=out,
/opt/conda/lib/python3.10/site-packages/numpy/lib/nanfunctions.py:1217: RuntimeWarning: All-NaN slice encountered
r, k = function_base._ureduce(a, func=_nanmedian, axis=axis, out=out,
CPU times: user 27.1 s, sys: 4.75 s, total: 31.8 s
Wall time: 5min 36s
# we turned ~2.6GiB of data into a 2.7MB GIF!
dask.utils.format_bytes(len(gif_img.data))
'2.68 MiB'
gif_img